The family Herpesviridae constitutes a large and diverse group of double-stranded DNA viruses found in a wide range of animal species, from invertebrates to humans. To date, nine distinct herpesviruses have been identified that cause lifelong infections in humans. These are collectively known as the Human Herpesviruses (HHV). Their biological and clinical significance is immense, ranging from common, relatively benign conditions like cold sores to severe, life-threatening diseases, including encephalitis, pneumonia, and multiple forms of cancer, particularly in immunocompromised individuals.
The defining characteristic of all herpesviruses is their ability to establish a lifelong, latent infection within their host. Following a primary lytic infection, the virus can enter a dormant state in specific cell types, only to reactivate later under certain stimuli. This complex lifecycle of lytic replication, latency, and reactivation is the central challenge in developing curative therapies and effective vaccines. A profound understanding of these fundamental viral processes is paramount for advancing novel therapeutics, a core focus of the preclinical research solutions provided by CD BioSciences. Our mission is to empower biotech and pharmaceutical partners by providing the specialized tools, assays, and models necessary to dissect HHV biology and accelerate drug discovery pipelines.
Introduction to Herpesviridae
The family Herpesviridae is formally classified by the International Committee on Taxonomy of Viruses (ICTV). These viruses are ubiquitous in the human population, with seroprevalence for some members, like Epstein-Barr Virus (EBV), exceeding 90% in adults worldwide. While often asymptomatic, their capacity to cause recurrent disease and their association with oncogenesis make them a persistent global health concern and a high-priority target for virological research and therapeutic development.
Classification: Alpha-, Beta-, and Gamma-herpesviruses
The Herpesviridae family is subdivided into three subfamilies—Alphaherpesvirinae, Betaherpesvirinae, and Gammaherpesvirinae—based on their distinct biological properties, such as host range, duration of the reproductive cycle, and the specific cell types in which they establish latency.
Alpha-herpesviruses (Alphaherpesvirinae)
Alpha-herpesviruses are characterized by a relatively broad host range, a short and efficient lytic replication cycle that rapidly destroys the host cell, and the ability to establish latency primarily in the sensory neurons of the host.
Members
Herpes Simplex Virus 1 (HSV-1), Herpes Simplex Virus 2 (HSV-2), and Varicella-Zoster Virus (VZV; HHV-3).
Biological Hallmarks
Rapid replication leads to visible cytopathic effects in cell culture. Their latency in non-dividing neurons allows them to persist for the lifetime of the host, evading immune surveillance. Reactivation, often triggered by stress or immunosuppression, results in viral transport back to mucosal or cutaneous sites, causing recurrent lesions.
Beta-herpesviruses (Betaherpesvirinae)
Beta-herpesviruses have a more restricted host range and a significantly longer replication cycle compared to their alpha-herpesvirus cousins. They are known for their ability to cause cellular enlargement (cytomegalia) and establish latency in a variety of cell types, including hematopoietic stem cells, monocytes, and endothelial cells.
Members
Human Cytomegalovirus (CMV; HHV-5), Human Herpesvirus 6A and 6B (HHV-6A, HHV-6B), and Human Herpesvirus 7 (HHV-7).
Biological Hallmarks
Slow, prolonged replication cycle. Latency in myeloid lineage cells acts as a systemic reservoir for dissemination and reactivation, posing a significant threat to transplant recipients and other immunocompromised patients.
Gamma-herpesviruses (Gammaherpesvirinae)
Gamma-herpesviruses are distinguished by their very narrow host range, typically specific to the family or order of the host. They replicate in epithelial and fibroblastic cells but uniquely establish latency within lymphoid cells (B or T lymphocytes). This tropism for lymphocytes is directly linked to their potent oncogenic potential.
Members
Epstein-Barr Virus (EBV; HHV-4) and Kaposi's Sarcoma-associated Herpesvirus (KSHV; HHV-8).
Biological Hallmarks
Their ability to establish latency in B-lymphocytes (EBV) or endothelial and B cells (KSHV) and to express viral genes that manipulate host cell growth and survival pathways underpins their association with cancers like Burkitt's lymphoma, nasopharyngeal carcinoma, and Kaposi's sarcoma.
Common Features: Structure, Genome, and Replication Cycle
Despite their subfamily-specific differences, all human herpesviruses share a conserved, multi-layered structure and a common strategy for replication.
- Virion Structure
The mature herpesvirus virion is a sophisticated, roughly spherical particle, approximately 200 nm in diameter, composed of four distinct architectural components:
Core
The innermost part, containing the large, linear double-stranded DNA (dsDNA) genome.
Capsid
A protein shell with icosahedral symmetry (T=16) that encases and protects the viral genome. It is composed of 162 capsomeres.
Tegument
An amorphous protein layer situated between the capsid and the envelope. This unique compartment is not a rigid structure but a complex assortment of dozens of viral proteins. These tegument proteins are critical for the earliest stages of infection, playing roles in shutting down host protein synthesis, evading intrinsic immunity, and commandeering cellular machinery to initiate viral gene expression.
Envelope
A lipid bilayer derived from host cell membranes (often the trans-Golgi network or nuclear membrane) and studded with numerous viral glycoproteins. These glycoproteins are essential for viral attachment to host cells and fusion between the viral envelope and a host cell membrane to deliver the capsid and tegument into the cytoplasm.
- Viral Genome
The HHV genome is a linear dsDNA molecule ranging from approximately 125 to 240 kilobase pairs (kbp), making it one of the largest among human viruses. A common organizational feature is the presence of unique long (UL) and unique short (US) regions, flanked by terminal and internal inverted repeat sequences. These repeats facilitate genomic isomerization and are involved in replication and packaging. The genome encodes between 70 and over 200 proteins, which are responsible for viral DNA replication, virion assembly, immune evasion, and the manipulation of host cellular processes.
- The Lytic Replication Cycle
Lytic replication is a highly regulated and temporally controlled cascade of gene expression that results in the production of new progeny virions and, ultimately, the death of the host cell.
1. Attachment and Entry: The cycle begins when viral glycoproteins on the envelope bind to specific receptors on the host cell surface, triggering membrane fusion and the release of the capsid and tegument into the cytoplasm.
2. Nuclear Targeting and Genome Injection: The capsid is transported along microtubules to the nucleus, where it docks at a nuclear pore complex and injects the viral DNA into the nucleoplasm.
3. Temporal Gene Expression: Viral gene expression proceeds in a coordinated cascade, orchestrated by both viral and host transcription factors.
- Immediate-Early (IE or alpha) Genes: Transcribed first, these genes encode regulatory proteins that take control of the host cell and activate the transcription of early genes.
- Early (E or beta) Genes: These encode proteins required for viral DNA replication, such as the viral DNA polymerase and helicase-primase complex. These enzymes are key targets for most licensed antiviral drugs.
- Late (L or gamma) Genes: Transcribed after the onset of viral DNA replication, these genes primarily encode the structural proteins needed to build new virions, such as capsid and tegument proteins, as well as glycoproteins.
4. DNA Replication & Assembly: The viral genome is replicated in the nucleus. Newly synthesized genomes are then packaged into pre-assembled capsids.
5. Egress: The filled capsids exit the nucleus through a complex process of primary envelopment at the inner nuclear membrane and de-envelopment at the outer nuclear membrane. They acquire their final tegument and envelope in the cytoplasm before being released from the cell via exocytosis.
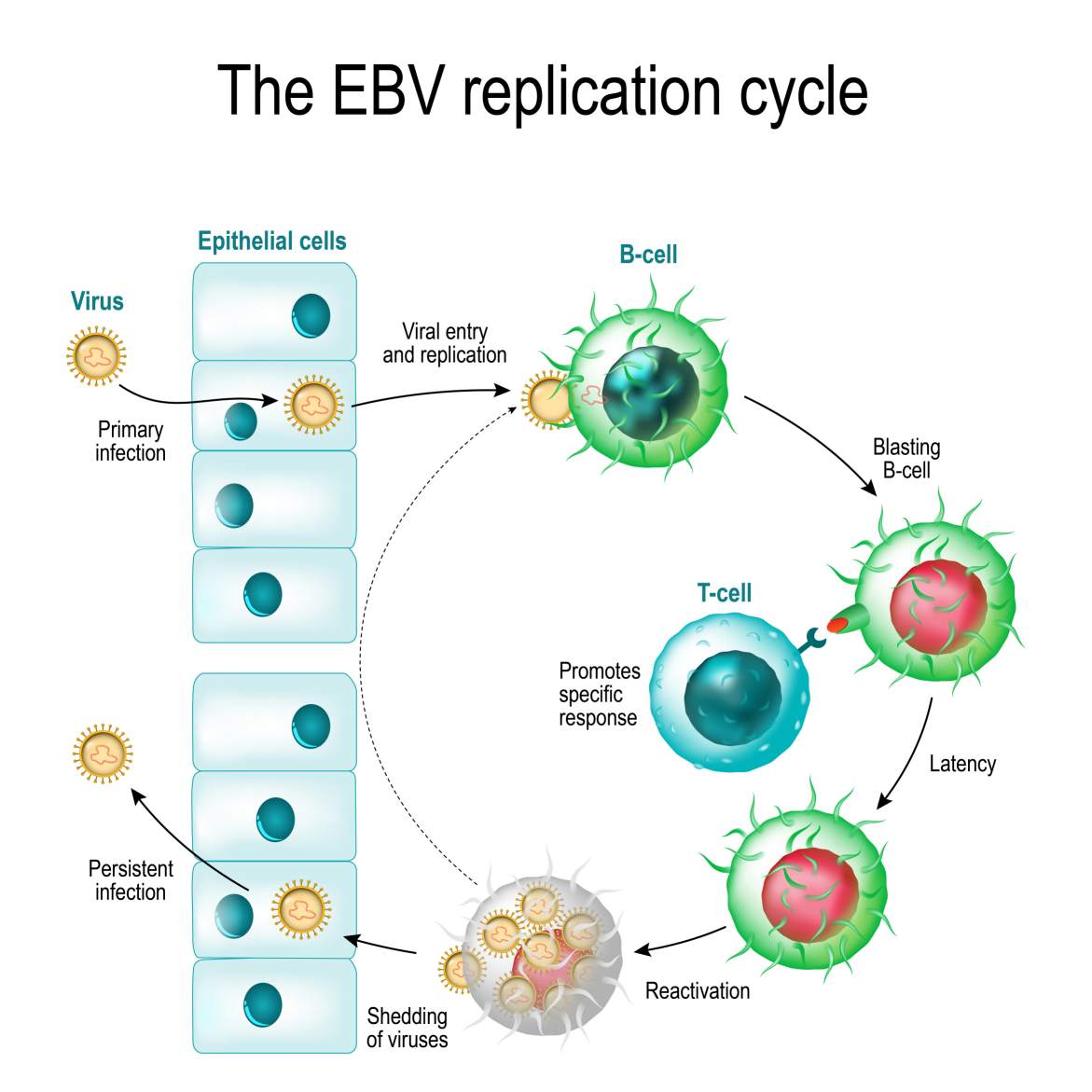
At CD BioSciences, we offer a suite of services to interrogate this cycle, from high-throughput screening assays targeting viral polymerases to advanced imaging of virion assembly and trafficking, supporting the preclinical evaluation of novel antiviral compounds.
The Hallmarks of Latency and Reactivation
The capacity for latency is the biological cornerstone of the Herpesviridae family's evolutionary success. It is a state of dynamic equilibrium between the virus and its host.
- Establishing and Maintaining Latency
Latency is a non-productive, reversible state of infection. Following primary infection, instead of undergoing lytic replication, the virus can enter a quiescent state in a specific reservoir of long-lived cells. During latency:
The viral genome persists as a circular, extrachromosomal plasmid (an episome) in the host cell nucleus.
The lytic gene expression cascade is almost completely silenced.
A very limited set of viral genes, known as latency-associated transcripts (LATs), are expressed. These genes play crucial roles in maintaining the latent state, protecting the infected cell from apoptosis, and helping the virus evade immune detection. For the oncogenic gamma-herpesviruses, latency proteins like EBV's EBNA1 and KSHV's LANA are essential for both maintaining the viral episome and driving cell proliferation.
- Reactivation: Awakening the Virus
Reactivation is the process by which a latent virus re-enters the lytic replication cycle. This switch is often triggered by physiological or external stimuli, including:
Host immunosuppression (e.g., due to medication, other infections like HIV, or aging)
Physical or psychological stress
UV radiation exposure
Hormonal changes
Trauma or inflammation
Upon receiving a reactivation signal, the repression of lytic genes is lifted, the IE gene cascade is initiated, and the full replication program resumes. This process is responsible for recurrent disease (e.g., cold sores, genital herpes, shingles) and is the primary route for viral shedding and transmission to new hosts.
Modeling latency and reactivation in vitro and in vivo represents a significant challenge and a critical frontier in HHV research. CD BioSciences is dedicated to this challenge, providing specialized cell-based latency models and animal models to help our partners study these complex processes and evaluate novel latency-disrupting or latency-maintaining therapeutic strategies for preclinical development. All our products and services are strictly for research use only (RUO).
References
- Griffin, B. D., Damania, B., & Knipe, D. M. (2020). Herpesviruses. In Fields Virology: Emerging Viruses (7th ed., pp. 249-284). Wolters Kluwer.
- Cohen, J. I. (2020). Herpesvirus Latency. Journal of Clinical Investigation, 130(7), 3361–3369. https://doi.org/10.1172/JCI136225
- Heldwein, E. E., & Krummenacher, C. (2008). Herpesvirus entry: a tale of two membranes. Current Opinion in Virology, 1(1), 39-49. https://doi.org/10.1016/j.coviro.2011.05.005
- Kieff, E., & Rickinson, A. B. (2007). Epstein-Barr Virus and Its Replication. In D. M. Knipe & P. M. Howley (Eds.), Fields Virology (5th ed., Vol. 2, pp. 2603–2654). Lippincott Williams & Wilkins.